Y.-Y. Liu and A.-L. Barabasi
Control Principles of Complex Systems
Review of Modern Physics 88: 3, 035006-035064 (2016)
Read the abstract
A reflection of our ultimate understanding of a complex system is our ability to control its behavior. Typically, control has multiple prerequisites: it requires an accurate map of the network that governs the interactions between the system’s components, a quantitative description of the dynamical laws that govern the temporal behavior of each component, and an ability to influence the state and temporal behavior of a selected subset of the components. With deep roots in dynamical systems and control theory, notions of control and controllability have taken a new life recently in the study of complex networks, inspiring several fundamental questions: What are the control principles of complex systems? How do networks organize themselves to balance control with functionality? To address these questions here recent advances on the controllability and the control of complex networks are reviewed, exploring the intricate interplay between the network topology and dynamical laws. The pertinent mathematical results are matched with empirical findings and applications. Uncovering the control principles of complex systems can help us explore and ultimately understand the fundamental laws that govern their behavior.
pdf
Article
jpgFigure 1
jpgFigure 2
jpgFigure 3
jpgFigure 4
jpgFigure 5
jpgFigure 6
jpgFigure 7
jpgFigure 8
jpgFigure 9
jpgFigure 10
jpgFigure 11
jpgFigure 12
jpgFigure 13
jpgFigure 14
jpgFigure 15
jpgFigure 16
jpgFigure 17
jpgFigure 18
jpgFigure 19
jpgFigure 20
jpgFigure 21
jpgFigure 22
jpgFigure 23
jpgFigure 24
jpgFigure 25
jpgFigure 26
jpgFigure 27
jpgFigure 28
jpgFigure 29
jpgFigure 30
jpgFigure 31
jpgFigure 32
jpgFigure 33
jpgFigure 34
jpgFigure 35
jpgFigure 36
jpgFigure 37
jpgFigure 38
jpgFigure 39
jpgFigure 40
jpgFigure 41
I. A. Kovács, A.-L. Barabási
Destruction perfected
Nature (News & Views) 524, 38-39 (2015)
Read the abstract
Pinpointing the nodes whose removal most effectively disrupts a network has become a lot easier with the development of an efficient algorithm. Potential applications might include cybersecurity and disease control. See Letter p.65, by F. Morone and H. A. Makse (Supplementary 1).

Albert-László Barabási
The network takeover
Nature Physics 8, 14-16 (2012)
Read the abstract
Reductionism, as a paradigm, is expired, and complexity, as a field, is tired. Data-based mathematical models of complex systems are offering a fresh perspective, rapidly developing into a new discipline: network science.
M. Vidal, M. E. Cusick, A.-L. Barabasi
Interactome Networks and Human Disease
Cell 144, 986-995 (2011)
Read the abstract
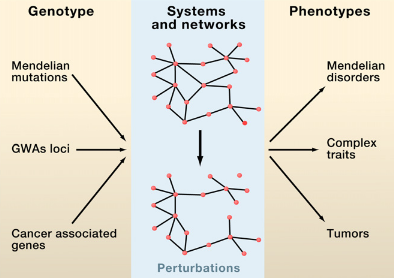
Complex biological systems and cellular networks may underlie most genotype to phenotype relationships. Here, we review basic concepts in network biology, discussing different types of interactome networks and the insights that can come from analyzing them. We elaborate on why interactome networks are important to consider in biology, how they can be mapped and integratedwith each other, what global properties are starting to emerge from interactome network models, and how these properties may relate to human disease.
A.-L. Barabási, N. Gulbahce, J. Loscalzo
Network medicine: a network-based approach to human disease
Nature Reviews Genetics 12, 56-68 (2011)
Read the abstract
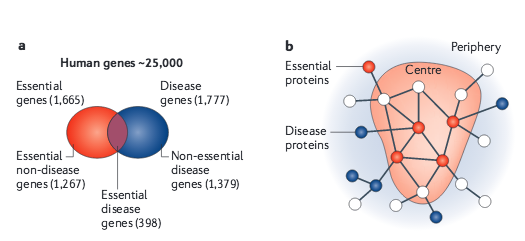
Given the functional interdependencies between the molecular components in a human cell, a disease is rarely a consequence of an abnormality in a single gene, but reflects the perturbations of the complex intracellular and intercellular network that links tissue and organ systems. The emerging tools of network medicine offer a platform to explore systematically not only the molecular complexity of a particular disease, leading to the identification of disease modules and pathways, but also the molecular relationships among apparently distinct (patho)phenotypes. Advances in this direction are essential for identifying new disease genes, for uncovering the biological significance of disease-associated mutations identified by genome-wide association studies and full-genome sequencing, and for identifying drug targets and biomarkers for complex diseases.
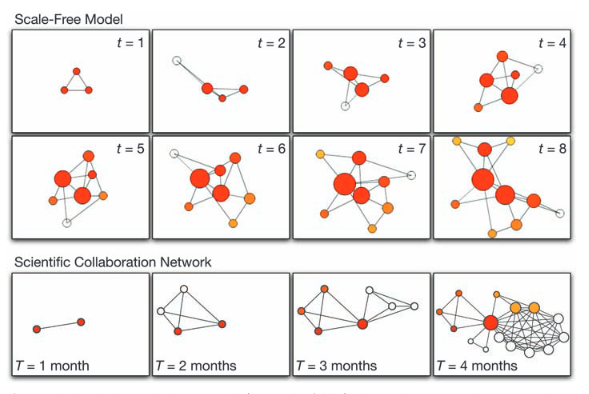
A.-L. Barabási
Scale-Free Networks: A Decade and Beyond
Science 325, 412-413 (2009)
Read the abstract
For decades, we tacitly assumed that the components of such complex systems as the cell, the society, or the Internet are randomly wired together. In the past decade, an avalanche of research has shown that many real networks, independent of their age, function, and scope, converge to similar architectures, a universality that allowed researchers from different disciplines to embrace network theory as a common paradigm. The decade-old discovery of scale-free networks was one of those events that had helped catalyze the emergence of network science, a new research field with its distinct set of challenges and accomplishments.
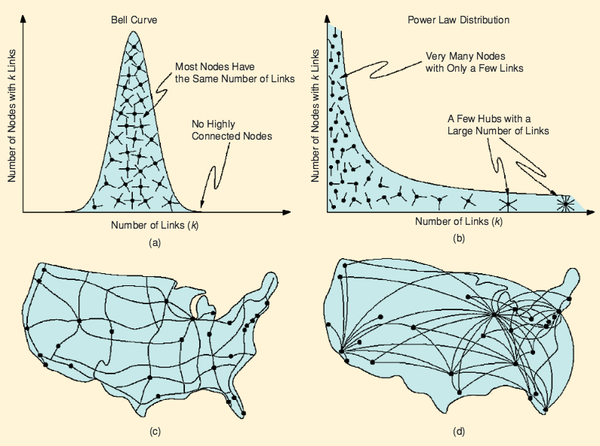
A.-L. Barabási
The architecture of complexity
IEEE Control Systems Magazine 27:4, 33-42 (2007)
Read the abstract
We are surrounded by complex systems, from cells made of thousands of molecules to society, a collection of billions of interacting individuals. These systems display signatures of order and self-organization. Understanding and quantifying this complexity is a grand challenge for science. Kinetic theory, developed at the end of the 19th century, shows that the measurable properties of gases, from pressure to temperature, can be reduced to the random motion of atoms and molecules. In the 1960s and 1970s, researchers developed systematic approaches to quantifying the transition from disorder to order in material systems such as magnets and liquids. Chaos theory dominated the quest to understand complex behavior in the 1980s with the message that unpredictable behavior can emerge from the nonlinear interactions of a few components. The 1990s was the decade of fractals, quantifying the geometry of patterns emerging in self-organized systems, from leaves to snowflakes.
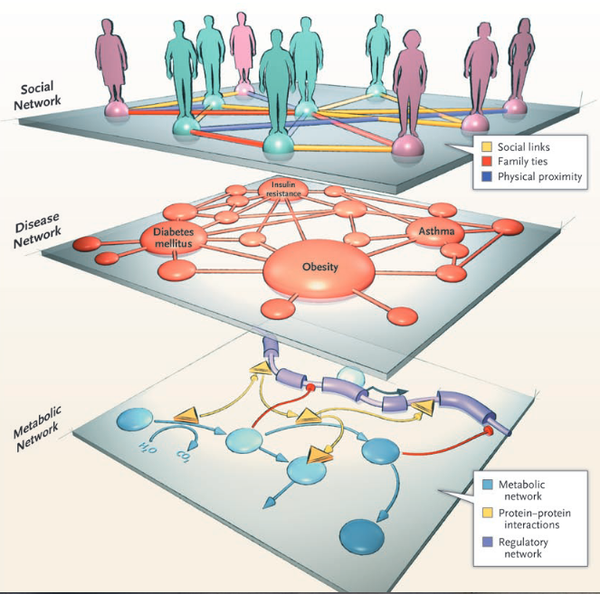
A.L. Barabási
Network Medicine — From Obesity to the “Diseasome”
New England Journal of Medicine 357, 404-407 (2007)
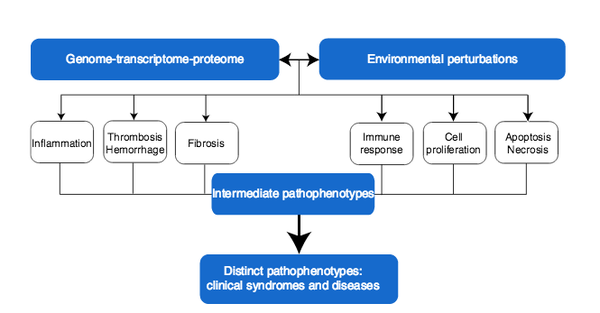
J. Loscalzo, I. Kohane, A.-L. Barabási
Human disease classification in the postgenomic era: A complex systems approach to human pathobiology
Molecular Systems Biology 3:124, 1-11 (2007)
Read the abstract
Contemporary classification of human disease derives from observational correlation between pathological analysis and clinical syndromes. Characterizing disease in this way established a nosology that has served clinicians well to the current time, and depends on observational skills and simple laboratory tools to define the syndromic phenotype. Yet, this time-honored diagnostic strategy has significant shortcomings that reflect both a lack of sensitivity in identifying preclinical disease, and a lack of specificity in defining disease unequivocally. In this paper, we focus on the latter limitation, viewing it as a reflection both of the different clinical presentations of many diseases (variable phenotypic expression), and of the excessive reliance on Cartesian reductionism in establishing diagnoses. The purpose of this perspective is to provide a logical basis for a new approach to classifying human disease that uses.
A.-L. Barabási
Taming complexity
Nature Physics 1, 68-70 (2005)
Read the abstract
The science of networks is experiencing a boom. But despite the necessary multidisciplinary approach to tackle the theory of complexity, scientists remain largely compartmentalized in their separate disciplines. Can they find a common voice?

A.-L. Barabási, E. Bonabeau
Scale-Free Networks
Scientific American 288, 50-59 (2003)
Read the abstract
Scientists have recently discovered that various complex systems have an underlying architecture governed by shared organizing principies. This insight has important implications for a host of applications, from drug development to Internet security.
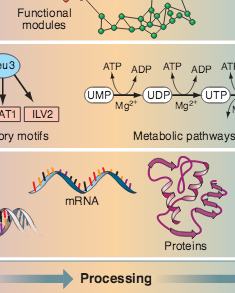
Z. N. Oltvai, A.-L. Barabási
Life’s complexity pyramid
Science 298, 763-764 (2002)
Read the abstract
Cells and microorganisms have an impressive capacity for adjusting their intracellular machinery in response to changes in their environment, food availability, and developmental state. Add to this an amazing ability to correct internal errors — battling the effects of such mistakes as mutations or misfolded proteins — and we arrive at a major issue of contemporary cell biology: our need to comprehend the staggering complexity, versatility, and robustness of living systems. Although molecular biology offers many spectacular successes, it is clear that the detailed inventory of genes, proteins, and metabolites is not sufficient to understand the cell’s complexity (1). As demonstrated by two papers in this issue—Lee et al. (2) on page 799 and Milo et al. (3) on page 824—viewing the cell as a network of genes and proteins offers a viable strategy for addressing the complexity of living systems.
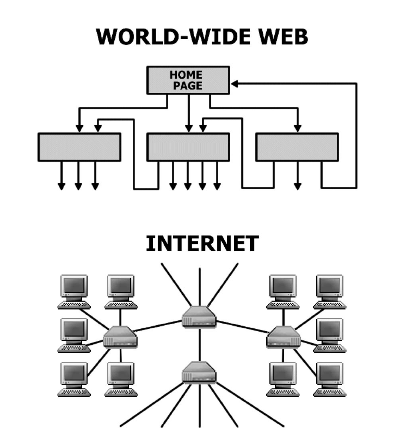
R. Albert, A.-L. Barabási
Statistical mechanics of complex networks
Reviews of Modern Physics 74, 47-97 (2002)
Read the abstract
Complex networks describe a wide range of systems in nature and society. Frequently cited examples include the cell, a network of chemicals linked by chemical reactions, and the Internet, a network of routers and computers connected by physical links. While traditionally these systems have been modeled as random graphs, it is increasingly recognized that the topology and evolution of real networks are governed by robust organizing principles. This article reviews the recent advances in the field of complex networks, focusing on the statistical mechanics of network topology and dynamics. After reviewing the empirical data that motivated the recent interest in networks, the authors discuss the main models and analytical tools, covering random graphs, small-world and scale-free networks, the emerging theory of evolving networks, and the interplay between topology and the network’s robustness against failures and attacks.

A.-L. Barabási
The physics of the web
Physics World 14, 33-38 (2001)
Read the abstract
Statistical mechanics is offering new insights into the structure and dynamics of the Internet, the World Wide Web and other complex interacting systems. TH E INTERNET appears to have taken on a life of its own ever since the National Science foundation in the US gave up stewardship of the network in 1995. New lines and routers are added continually by thousands of companies, none of which require permission from anybody to do so, and none of which are obliged to report their activity. This uncontrolled and decentralized growth has turned network designers into scientific explorers. All previous Internet-related research concentrated on designing better protocols and faster components. More recently, an increasing number of scientists have begun to ask an unexpected question: what exactly did we create?'
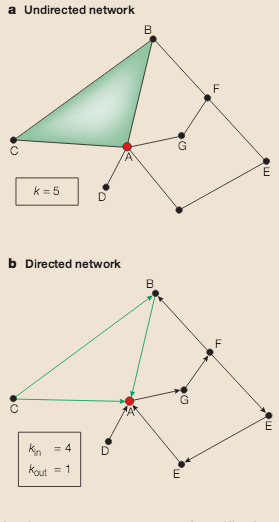
A.-L. Barabási, Z. N. Oltvai
Network biology: understanding the cell’s functional organization
Nature Reviews Genetics 5, 101-113 (2004)
Read the abstract
A key aim of postgenomic biomedical research is to systematically catalogue all molecules and their interactions within a living cell. There is a clear need to understand how these molecules and the interactions between them determine the function of this enormously complex machinery, both in isolation and when surrounded by other cells. Rapid advances in network biology indicate that cellular networks are governed by universal laws and offer a new conceptual framework that could potentially revolutionize our view of biology and disease pathologies in the twenty-first century.