A.-L. Barabási, R. Albert
Emergence of scaling in random networks
Science 286, 509–512 (1999)
Read the abstract
Systems as diverse as genetic networks or the World Wide Web are best described as networks with complex topology. A common property of many large networks is that the vertex connectivities follow a scale-free power-law distribution. This feature was found to be a consequence of two generic mechanisms: (i) networks expand continuously by the addition of new vertices, and (ii) new vertices attach preferentially to sites that are already well connected. A model based on these two ingredients reproduces the observed stationary scale-free distributions, which indicates that the development of large networks is governed by robust self-organizing phenomena that go beyond the particulars of the individual systems.
R. Albert, A.-L. Barabási
Statistical mechanics of complex networks
Reviews of Modern Physics 74, 47-97 (2002)
Read the abstract
Complex networks describe a wide range of systems in nature and society. Frequently cited examples include the cell, a network of chemicals linked by chemical reactions, and the Internet, a network of routers and computers connected by physical links. While traditionally these systems have been modeled as random graphs, it is increasingly recognized that the topology and evolution of real networks are governed by robust organizing principles. This article reviews the recent advances in the field of complex networks, focusing on the statistical mechanics of network topology and dynamics. After reviewing the empirical data that motivated the recent interest in networks, the authors discuss the main models and analytical tools, covering random graphs, small-world and scale-free networks, the emerging theory of evolving networks, and the interplay between topology and the network’s robustness against failures and attacks.
H. Jeong, B. Tombor, R. Albert, Z. N. Oltvai, A.-L. Barabási
The large-scale organization of metabolic networks
Nature 407, 651–655 (2000)
Read the abstract
Here we present a systematic comparative mathematical analysis of the metabolic networks of 43 organisms representing all three domains of life.We show that, despite significant variation in their individual constituents and pathways, these metabolic networks have the same topological scaling properties and show striking similarities to the inherent organization of complex non-biological systems. This may indicate that metabolic organization is not only identical for all living organisms, but also complies with the design principles of robust and error-tolerant scale-free networks, and may represent a common blueprint for the large-scale organization of interactions among all cellular constituents.
R. Albert, H. Jeong, A.-L. Barabási
Error and attack tolerance of complex networks
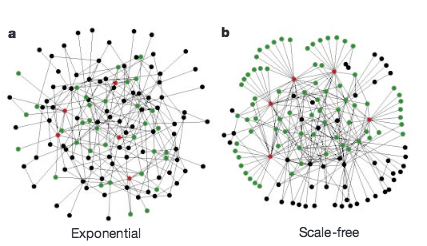
Nature 406, 378–482 (2000)
Read the abstract
Here we demonstrate that error tolerance is not shared by all redundant systems: it is displayed only by a class of inhomogeneouslywired networks,called scale-free networks, which include theWorld-WideWeb, the Internet, social networks and cells. We find that such networks display an unexpected degree of robustness, the ability of their nodes to communicate being unaffected even by unrealistically high failure rates.However, error tolerance comes at a high price in that these networks are extremely vulnerable to attacks (that is, to the selection and removal of a few nodes that play a vital role in maintaining the network’s connectivity). Such error tolerance and attack vulnerability are generic properties of communication networks.
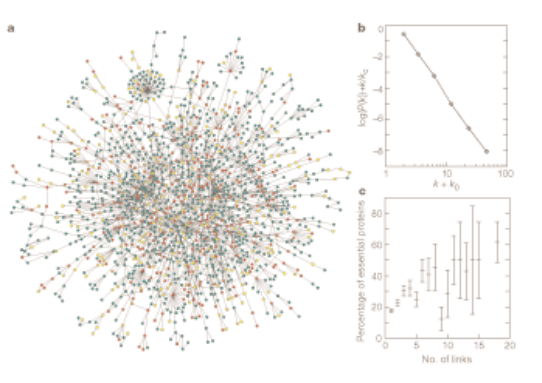
H. Jeong, S. P. Mason, A.-L. Barabási, Z. N. Oltvai
Lethality and centrality in protein networks
Nature 411, 41-42 (2001)
Read the abstract
The most highly connected proteins in the cell are the most important for its survival. Proteins are traditionally identified on the basis of their individual actions as catalysts, signalling molecules, or building blocks in cells and microorganisms. But our post-genomic view is expanding the protein’s role into an element in a network of protein–protein interactions as well, in which it has a contextual or cellular function within functional modules1,2. Here we provide quantitative support for this idea by demonstrating that the phenotypic consequence of a single gene deletion in the yeast Saccharomyces cerevisiae is affected to a large extent by the topological position of its protein product in the complex hierarchical web of molecular interactions.
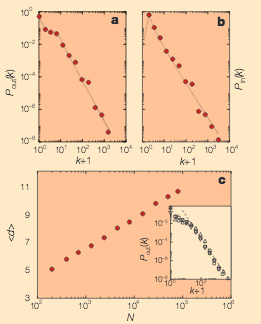
R. Albert, H. Jeong, A.-L. Barabási
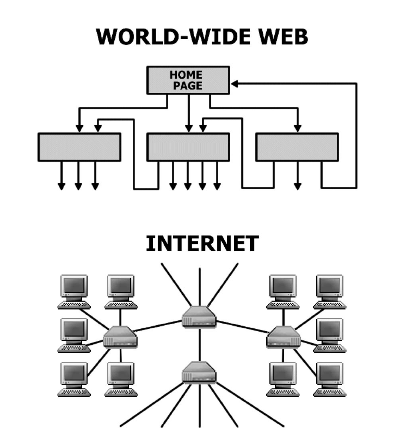
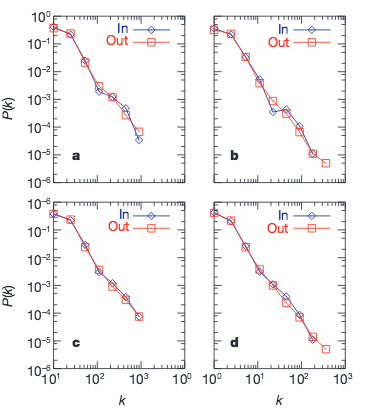
Diameter of the world wide web
Nature 401, 130-131 (1999)
Read the abstract
Despite its increasing role in communication, the World-Wide Web remains uncontrolled: any individual or institution can create a website with any number of documents and links. This unregulated growth leads to a huge and complex web, which becomes a large directed graph whose vertices are documents and whose edges are links (URLs) that point from one document to another. The topology of this graph determines the web’s connectivity and consequently how effectively we can locate information on it.
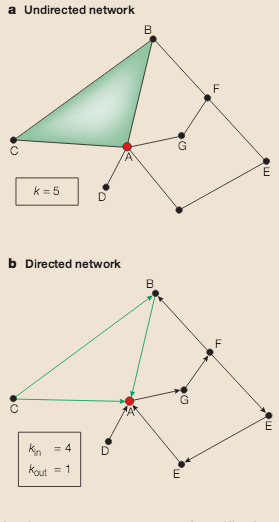
A.-L. Barabási, Z. N. Oltvai
Network biology: understanding the cell’s functional organization
Nature Reviews Genetics 5, 101-113 (2004)
Read the abstract
A key aim of postgenomic biomedical research is to systematically catalogue all molecules and their interactions within a living cell. There is a clear need to understand how these molecules and the interactions between them determine the function of this enormously complex machinery, both in isolation and when surrounded by other cells. Rapid advances in network biology indicate that cellular networks are governed by universal laws and offer a new conceptual framework that could potentially revolutionize our view of biology and disease pathologies in the twenty-first century.
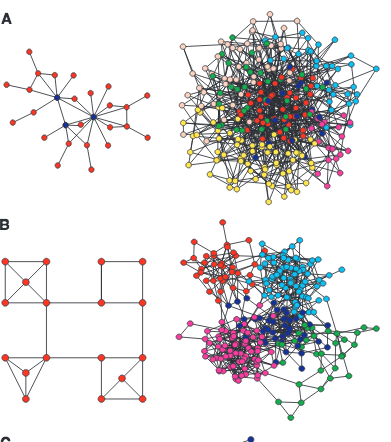
E. Ravasz, A. L. Somera, D. A. Mongru, Z. N. Oltvai, A.-L. Barabási
Hierarchical organization of modularity in metabolic networks
Science 297, 1551-1555 (2002)
Read the abstract
Spatially or chemically isolated functional modules composed of several cellular components and carrying discrete functions are considered fundamental building blocks of cellular organization, but their presence in highly integrated biochemical networks lacks quantitative support. Here, we show that the metabolic networks of 43 distinct organisms are organized into many small, highly connected topologic modules that combine in a hierarchical manner into larger, less cohesive units, with their number and degree of clustering following a power law. Within Escherichia coli, the uncovered hierarchical modularity closely overlaps with known metabolic functions. The identified network architecture may be generic to system-level cellular organization.
A.-L. Barabási, R. Albert, H. Jeong
Mean-field theory for scale-free random networks
Physica A 272, 173–187 (1999)
Read the abstract
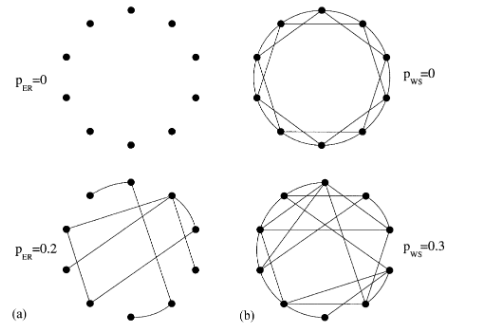
Random networks with complex topology are common in Nature, describing systems as diverse as the world wide web or social and business networks. Recently, it has been demonstrated that most large networks for which topological information is available display scale-free features. Here we study the scaling properties of the recently introduced scale-free model, that can account for the observed power-law distribution of the connectivities. We develop a mean-feld method to predict the growth dynamics of the individual vertices, and use this to calculate analytically the connectivity distribution and the scaling exponents. The mean-feld method can be used to address the properties of two variants of the scale-free model, that do not display power-law scaling.
R. Albert, A.-L. Barabási
Topology of evolving networks: local events and universality
Physical Review Letters 85, 5234-5237 (2000)
Read the abstract
Networks grow and evolve by local events, such as the addition of new nodes and links, or rewiring of links from one node to another. We show that depending on the frequency of these processes two topologically different networks can emerge, the connectivity distribution following either a generalized power law or an exponential. We propose a continuum theory that predicts these two regimes as well as the scaling function and the exponents, in good agreement with numerical results. Finally, we use the obtained predictions to fit the connectivity distribution of the network describing the professional links between movie actors.